Sleep apnea syndrome detection and classification of severity level from ECG and SpO2 signals
Introduction
Sleep apnea-hypopnea syndrome (SAHS) is a serious respiratory disorder defined as an interruption of normal respiration during sleep, which involves involuntary cessation of breathing that occurs while a person is a sleep. Loud disruptive snoring, brief repeated episodes of cessation of breathing during sleep, and daytime sleepiness characterizes SAHS. Apnea is defined as the airflow cessation for a time of at least 10 seconds (1-3). Hypopnea exists when either there is a 30% or 50% decrease in airflow for a minimum of 10 seconds duration (2,4).
Frequent apnea results in periodic lowering of blood oxygen concentrations and this forces the body to send signals to the lungs and chest to breathe harder. Airway size and function, lung tissue, lung’s blood supply, and breathing muscles (chest, diaphragm, and throat) among the determining factors for irregular breathing. Most of lung’s activities are controlled by the brain. When we are without sleep, the brain usually sends signals to the muscles of the chest and the throat, maintaining normal breathing.
The American Thoracic Society (ATS) indicated that the prevalence of sleep apnea affects more than 936 million people around the globe, with disorder severity ranges from mild to severe (5). Even though the rate of obstructive sleep apnea (OSA) is being reported to be 1% to 4% by various studies, the prevalence of OSA ranged from 0.1% to 13%. In the middle-aged population, 50% of men and 25% of women risk OSA from moderate to severe as defined by an apnea-hypopnea index (AHI) greater than or equal to 15 (6). Sleep apnea occurs in people of all ages. It may be most common in the elderly (7). Even though people with obesity have high risk of sleep apnea disorder, thin people may also have sleep apnea. Most of the symptoms of sleep apnea are snoring, sleep disruption, and irregular breathing. The irregular sleep disruptions also interfere with the brain’s normal sleep pattern inflicting arousals and reducing the amount of sleep time spent in deep sleep (8). This disrupts sleeping patterns. The breathing irregularities may also decrease the body’s oxygen level which leads to stress on the heart, and possibly contribute to high blood pressure, heart attacks or stroke. Sleep apnea may also lead to periodic drops in concentration of oxygen in the blood, causing intermittent hypoxia that simulates altered blood oxygen saturation (SpO2) profiles, and exhibits increased sympathetic nerve activity (SNA) and hypertension (8). The drop in blood oxygen level can be also the cause of hypertension which is a major clinical problem of sleep apnea patients (9).
Sleep apnea-hypopnea diagnosis decision is made using a complete Polysomnography (PSG) test at sleep laboratories, where an expert human observer is needed to work overnight. The outcome of diagnosis using PSG relies on the expert experience. The results obtained could also be subjective leading to misdiagnosis. In order to improve diagnosis efficiency, reduce diagnosis time, and ensure a more accurate diagnosis, a quantitative and objective method is required.
Much of the current sleep apnea researches cover comprehensive portable devices to offline systems for automatic detection of sleep apnea by analyzing different signals stored on PSG records (10-14). Statistical, Time and frequency domain features of different signals such as electrical activity in response to a nerve’s stimulation of the muscle [electromyogram (EMG)], the thorax and abdomen effort signals, acoustic speech signal, SpO2, electrical activity of the brain [electroencephalograph (EEG)], and electrical activity of the heart [electrocardiograph (ECG)] are commonly used in the detection (15,16).
Many studies show that detection of OSA can be performed by using ECG derived respiration signal and/or heart rate variability (HRV) from ECG signal. Fast Fourier transform and Wavelet packet decomposition based on support vector machine (SVM) was used for feature extraction of HRV, and a 90% sensitivity and 93.34% accuracy were obtained (15). In this study, only HRV features were used. However, many studies indicated that HRV can be affected by other cardiological diseases which may affect their result. In addition, it didn’t include severity detection and the accuracy achieved was low.
Few studies have been proposed based on the arterial SpO2 signal measured by SpO2 to obtain high-quality signal features in discriminating OSA (10,14,17). Three features of the SpO2 signal (delta index, central tendency measured with radius 0.5 (CTM 50) and oxygen desaturation index (ODI) were extracted and analyzed to evaluate sleep quality (10). Artificial neural network-based sleep apnea (SA) detection using blood SpO2 features obtained from SPO2 has been proposed by Mostafa et al. (17) and an accuracy of 97.7% was claimed. However, the system did not incorporate severity classification which is an important parameter for treatment type given to the patient.
Signals from either EEG and SpO2 or ECG and SpO2 has been also proposed in literature for automated diagnosis of SA. The relationship between periodic changes in the SpO2 profile and in the EEG pattern due to apnea events during the night was investigated (18) using spectral analyses of these two signals and a 91% sensitivity, 83.3% specificity and 88.5% accuracy in OSA diagnosis was reported. However, the system has low accuracy and severity detection was not included in their system. A system that used an artificial neural network-based detection of OSA from ECG and SpO2 in combination was also developed and tested using 12 features from ECG signal and 2 features from and an accuracy of 98.3%, a specificity of 100% and a sensitivity of 96.6% was reported (19). Even though the accuracy achieved was good severity classification has not been considered.
There are several challenging problems in sleep disorder diseases which have not yet been solved adequately and efficiently, including sleep apnea detection or diagnosis and apnea severity classification. The aim of this research work was to develop simple and accurate automatic detection and severity level classification of sleep apnea from ECG and SpO2 signal individually and in combination using machine learning.
Methods
In this research ECG and SpO2 signals in combination has been used for detection of sleep apnea syndrome and severity classification. The proposed method includes three main parts: signal preprocessing, features extraction based on statistical measures and features classification. Feature classification is implemented using SVMs. Figure 1 below demonstrates the general block diagram of the proposed method employed in this research.
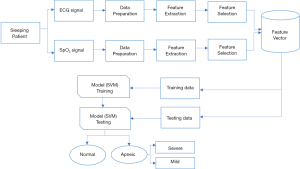
Data collection
A total of 400 ECG and SpO2 data have been collected from online dataset (PhysioNet database) and also 40 data were recorded from Hallelujah General Hospital sleep laboratory. The data which were collected from the PhysioNet apnea ECG database (20) contains a continuous digitalized ECG apnea annotated signals recorded from 70 patients with sampling frequency of 100 Hz. Among these, 8 records were accompanied by four additional SpO2 signals. The data recorded from Hallelujah General Hospital were recorded from 5 patients, 20 normal and 20 apneic, with sampling frequency of 200 Hz. This data was used for testing purpose. The signal acquired from PhysioNet database was resampled to 200 Hz for data uniformity. A written informed consent has been obtained from the participants prior to data collection. This research has been approved by Jimma Universitys institutional review board (IRB: 36/2019) and the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
ECG signal analysis
Preprocessing
ECG signal can be contaminated with different types of noise up on recording, which makes it difficult for the physician to interpret the signal. The most common noises or artifacts that encounter during ECG signal recording include baseline wander, power line interference, muscle noises, and electrode motion artifacts.
ECG signals are typically described by bandwidth of 0.5–100 Hz (21). Band pass filtering with cutoff frequency of these extreme values can remove unwanted signals in the preprocessing stage. Butterworth low pass and high pass filters with cutoff frequencies 100 and 0.5 Hz, respectively were used in the preprocessing stage to remove unwanted signals.
In ECG signal, the baseline wander is caused due to improper electrodes (electrode-skin impedance), patient’s movement and breathing (respiration). Wavelet transform were used for removing baseline wander from the ECG signal. A notch filter with frequency of 50 Hz were used to remove the power line interference. This is because the standard utility frequency of countries where the online data and local data were recorded (Germany and Ethiopia, respectively) is 50 Hz.
R-peak detection
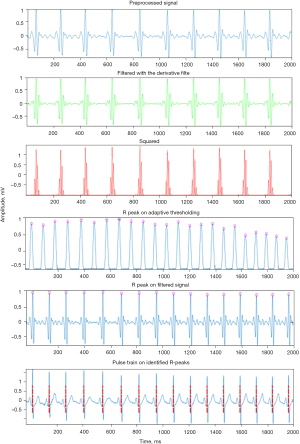
The QRS complex represents the electrical impulses spreads through ventricles and indicates ventricular depolarization. Not every but most of QRS complex contain Q, R, and S waves. In this paper, the Pan-Tompkins algorithm of (22), which uses three processing steps including linear digital filtering, nonlinear transformation, and decision rule algorithms was used for QRS complex detection. The performance of the method was evaluated in the presence of noise and a 99% accuracy was reported (20). The band pass filter with cutoff frequencies of 15 and 5 Hz selects the QRS and removes unwanted peaks; the derivate is used to get slope information; squaring function provides further attenuation of other ECG features leaving the QRS complexes as outstanding positive peaks in the signal regardless of their polarity in the original ECG recording, the moving window function produces a large amplitude pulse for every QRS and lower amplitude pulses for noise spikes, and finally the Adaptive thresholding selects peaks with large amplitudes (anticipated R peaks).
Feature extraction and selection
A total of 12 features were extracted for after R-peak detection. The Relief function has been used for feature selection (23). Relief calculates a feature score for every feature which may then be applied to rank and choose top-scoring options for feature selection. It relies on the identification of feature value variations between nearest-neighbor instance pairs. The function returns the rank and weights of predictors for the input data matrix. Table 1 illustrates feature extracted from ECG for apnea detection and classification of severity level along with their weight and predictor rank. The first 10 ranked features were selected for model training.
Table 1
| Rank | Features extracted | Predictor weight |
|---|---|---|
| 1 | A variance of the ratio of two consecutive RR intervals (Var. of R1/R2) | 0.08 |
| 2 | The root mean square of standard deviation measures or the square root of the mean of the sum of the squares of differences between adjacent RR-intervals (RMSSD) | 0.07 |
| 3 | Standard deviation of the differences between adjacent RR-intervals (SDSD) | 0.065 |
| 4 | Mean epoch of recording RR-interval (mean of RR) | 0.063 |
| 5 | The standard deviation of two consecutive RR intervals (Std. of R1/R2) | 0.06 |
| 6 | Median of RR-intervals (median of RR) | 0.055 |
| 7 | Mean of the ratio of two consecutive RR intervals (mean R1/R2) | 0.03 |
| 8 | Variance of the RR interval (Var. of RR) | 0.021 |
| 9 | Median of the Ratio of two consecutive RR intervals (median of R1/R2) | 0.017 |
| 10 | The standard deviation of the epoch and recording RR-interval (Std. of RR) | 0.015 |
| 11 | Percentage of NN50 (PP50) | 0.008 |
| 12 | Number of R peaks in ECG that differ more than 50 milliseconds (NN50) | −0.06 |
ECG, electrocardiograph; RR, successive R waves of ECG; RMSSD, root mean square of successive differences between adjacent RR-intervals; SDSD, standard deviation of the differences between adjacent RR-intervals.
SpO2 signal analysis
Pre-processing
The SpO2 signal data, which were recorded with the ECG signal simultaneously, were first separated and segmented in 1-minute intervals and processed off-line by an automated system, which was developed using MATLAB. The data acquired from PhysioNet database was already a labeled data for every minute. The data recorded from Hallelujah General Hospital was annotated with three experts. A minute is considered as apnea if apnea or hypopnea occurs for ten or more seconds. During preprocessing, a minute which has a blood oxygen level less than 50% are considered as artifacts and removed from the datasets.
Feature extraction and selection
A total of 10 features were extracted from the segmented SpO2 data. If no artifact was detected in the oximetry signal with in an epoch, then the following temporal SpO2 saturation features were calculated for each one-minute epoch using the sampled signal. The total features extracted are demonstrated Table 2. For feature selection, the same procedure which was used for ECG has been applied for SpO2 (using the Relief function). Features were ranked based on their feature importance weight. From the total of ten features extracted, the first seven features were selected for model training. The Mode of SpO2 value feature had a negative predictor important weight and the predictor ranked 8th and 9th also had a predictor importance weights less than 0.01 which are insignificant for classification.
Table 2
| Rank | Features extracted | Predictor weight |
|---|---|---|
| 1 | The mean SpO2 value (mean of SpO2) | 0.065 |
| 2 | The minimum SpO2 value (min of SpO2) | 0.06 |
| 3 | The Standard deviation of SpO2 value (Std. of SpO2) | 0.055 |
| 4 | The variance of the absolute difference between two successive SpO2 value (Var. of diff SpO2) | 0.031 |
| 5 | The maximum SpO2 value (max SpO2) | 0.022 |
| 6 | The mean of the absolute differences between successive SpO2 value (mean of diff SpO2) | 0.013 |
| 7 | Median of SpO2 value (median of SpO2) | 0.013 |
| 8 | The standard deviation of the absolute differences between successive SpO2 value (Std. of diff SpO2) | 0.005 |
| 9 | The variance of SpO2 value (Var. of SpO2) | 0.004 |
| 10 | The mode of SpO2 value (mode of SpO2) | −0.01 |
Classification
Different classifiers were assessed to select the best one, and SVM was selected and applied for our case. The SVM is classified under a supervised machine learning algorithm. It can be used for classification and regression purposes. The classification is performed by finding the hyperplane that differentiates the two classes very well (21). There may be many possible linear classifiers that can separate two classes, but the preferred one is that it maximizes the distance between it and the nearest data point of each class. This linear classifier is called the best separating hyperplane. The SVM algorithm builds a model that assigns new examples or test sets in one category or in the other category making it no probabilistic binary linear classifier. In SVM, P data is classified to which class it belongs, by points with a (P−1) dimensional hyperplane, which is called a linear classifier. A good separation between the 2 possible classes is achieved by building the greatest margin hyperplane. The margin maximizes the distance between the categories and therefore the nearest information of each category. The larger the margin is, the lower the generalization error of the classifier.
For our purpose, the SVM classifies the data as apnea event or normal event. Training and testing data were selected randomly. From the total of 400 data acquired from PhysioNet database, 280 data were used for training and the remaining 120 for testing. The remaining 40 data collected from Hallelujah General Hospital has been also used for further testing alone.
Severity classification has been also performed using the SVM classifier. A 1-hour raw ECG and SpO2 records with apnea or hypopnea events were used for this purpose. Then similar procedures used for the detection of sleep apnea algorithm including signal preprocessing and feature extraction were used for severity classification. For this purpose, 8 subjects of which approximately 8-hour signal was collected. From 6 subject 8 samples and from 2 subjects 7 samples, totally 62 samples were acquired by counting the number of apnea or hypopnea event occurrence in 1-hour signal. Then a signal which contains apneic event more than 30 per hour were considered as sever and a 1-hour signal which contain apneic event between 5 and 30 were annotated as not severe. From the total 62 data, 70% were used for training and the reaming 30% for testing.
Results
ECG signal analysis
ECG pre-processing and R-peak detection
The preprocessing steps involved in ECG signal include random noise removal by using a band pass filter, removing power line interference by using a notch filter and removing baseline wander by using wavelet denoising. Figure 2 shows the results of each algorithm used for R-peak detection including the band pass filter, derivative, squaring, and adaptive Thresholding.
ECG based apnea classification result
To test the proposed method 400 sample signals were used from PhysioNet, out of which 232 are apnea positive events and 168 are apnea negative events. From the total 400 samples, 70% (which is around 280 samples) are used for training purpose and the remaining 30% (which is around 120 samples) are used for testing purposes. From the total of 68 apnea signals, 67 were correctly predicted as apnea while 1 was predicted normal. Similarly, from a total of 52 normal signals, 47 were correctly predicted as normal while 5 were predicted as apnea. The results show that using SVM classifier an accuracy of 95%, the sensitivity of 98.5% and specificity 90.4% were achieved.
For comparison, the training was also conducted using K-Nearest Neighbors (KNN) and linear discriminant analysis (LDA) classifiers. An accuracy of 94.2%, sensitivity of 97.1% and specificity of 90.2% were achieved using KNN while for LDA a 93.3% accuracy, 98.5% sensitivity and 86.5% specificity was found. Our experimental results shows that SVM is superior than LDA and KNN in classification of apnea based on ECG signal.
Classification test result of ECG signal using data collected from Hallelujah General Hospital
The proposed model has been also tested by using data acquired from Hallelujah General Hospital. A total of 40 ECG data, 20 normal and the remaining 20 apneic, were used for additional testing purpose. Among these, 19 were classified as apnea and 17 as normal. Three normal labeled signals were misclassified as apnea and the reaming 1 apnea labeled signal was classified as normal. As a result, an accuracy, sensitivity and specificity of 90%, 95% and 85% has been achieved, respectively.
Apnea classification using SpO2 signal alone
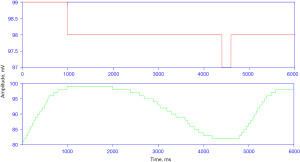
During preprocessing an epoch or a minute signal with SpO2 less than 50% has been removed from the data set as part of the noise removal step. Figure 3 shows the sample SpO2 signal for normal and apnea events.
A total of 120 SpO2 data, 68 apnea and 52 normal has been used for training the SVM classifier. Among these, all 68 apnea signals were classified as apnea with no misclassification while 2 normal labeled signals were misclassified as apnea from the total 52 normal signals. Accordingly, an accuracy of 98.3%, sensitivity of 100% and specificity of 96.2% has been found using the proposed classifier. The data was also trained using LDA (97% accuracy), KNN (95.8% accuracy) classifiers for comparison purpose and dominant performance was achieved by using the SVM classifier (98.3% accuracy). Additionally, A total of 40 SpO2 data: 20 normal and the remaining 20 apneic, collected from Hallelujah General Hospital, were used for testing purpose. Among these, 19 were correctly classified as apnea and 18 as normal. Two normal labeled signals were misclassified as apnea and the reaming 1 apnea labeled signal was misclassified as normal. Accordingly, an accuracy of 92.5%, sensitivity of 95% and Specificity of 90% has been achieved.
SpO2 and ECG feature combination classification test result
Ten features extracted from ECG and 7 features extracted from SpO2 were used to train the SVM model for apnea classification using the ECG and SpO2 data recorded simultaneously. Using the combined features of SpO2 and ECG by using SVM classifier, an accuracy of 99.1%, specificity of 98.1% and sensitivity of 100% were achieved. LDA and KNN classifiers were also trained using combined feature sets and SVM show dominance performance as demonstrated in Table 3.
Table 3
| Classifier | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| LDA | 97.4 | 100 | 98.3 |
| KNN | 97.4 | 100 | 98.3 |
| SVM | 100 | 98.1 | 99.1 |
KNN, k-nearest neighbors; LDA, linear discriminant analysis; SVM, support vector machine.
A classification test result using local data (Hallelujah General Hospital) resulted an accuracy of 97.5%, sensitivity of 100% and specificity of 95%. Using ECG and SpO2 signals simultaneously offers benefits over using either signal alone.
Severity classification
For severity classification, an accuracy of 77.8%, sensitivity of 57.1% and specificity of 90.9% were achieved using ECG signal alone. By using SpO2 signal alone an accuracy of 83.3%, sensitivity of 71.4% and specificity of 90.9% were achieved. Finally, by using the combination of the two signals we found an accuracy of 88.9%, sensitivity of 85.7% and specificity of 90.9%. The result achieved by the combination of the two signals indicated that using the two signals at the same time improves the overall performance of the proposed system (Table 4).
Table 4
| Signal | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|
| ECG | 57.1 | 90.9 | 77.8 |
| SpO2 | 71.4 | 90.9 | 83.3 |
| ECG and SpO2 | 85.7 | 90.9 | 88.9 |
ECG, electrocardiograph.
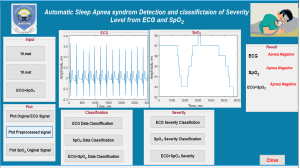
Graphical user interface (GUI) for apnea classification and severity level classification
A graphical interface has been developed for ease of use of the proposed method. It allows users to load ECG and SpO2 data, pre-process the signals, classify the signal and identify the severity level if it is apnea positive. Figure 4 demonstrates the GUI during apnea classification and severity level classification.
Discussion
The current gold standard sleep apnea detection device, PSG uses many sensors to acquire signals from the body. Reducing the number of signal recordings from PSG takes special importance in sleep studies because the use of many signal sensors may disturb the physiological sleep and affect the analysis. Using one signal alone or two signals in combination reduce the number of sensors and helps to decrease disturbances during physiological sleep and make the analysis simple. Automating sleep apnea diagnosis is also very important in reducing diagnosis errors that are brought through the current traditional manual diagnosis technique.
Previously sleep apnea detection has been proposed in literature based on ECG and SpO2 alone (10-12,15,16,24), and in a few cases also based on a combination of ECG and SpO2 (19,25). However, in most literature, either the data used for training and testing or the accuracy claimed is less which compromises the reliability of diagnosis. In addition, the severity of apnea, which is an important indicator of the level of sleep apnea that treatment is based on, has not been addressed in the literature proposed so far.
Sleep apnea syndrome detection and classification of severity level based on a simultaneously recorded ECG and SpO2 signals, individually and in combination has been evaluated in this thesis. Our method also incorporates a user-friendly GUI. The ECG and SpO2 signals carry important information related to the cardiovascular function as well as respiration. These signals present interesting characteristics that can be used to detect apneic events. The study showed that the ECG RR interval features and SpO2 features in combination improve the performance of sleep apnea diagnosis. Both linear and nonlinear features are selected in our proposed classifier that integrates SpO2 and ECG.
By using ECG signal features alone, we have obtained inaccuracy of 95% and by using SpO2 signal features alone an accuracy of 98.3% has been obtained. Our method demonstrated that better detection accuracy has been obtained using SpO2 alone compared to ECG alone. The result indicates features obtained from SpO2 seems to be highly significant indicators of SAHA compared to features obtained from RR interval of the ECG signal. Using a combination of features of simultaneously recorded ECG and SpO2 signals improve the accuracy (99.1%). This shows that the performance of the sleep apnea detection method is better in the hybrid compared to using each signal alone. Sleep apnea severity classification has been also incorporated in our method. Using the hybrid method (SpO2 + ECG) 88.9% accuracy has been achieved for severity classification.
A method to detect whether a patient is OSA positive or OSA negative from the SpO2 signal was proposed (10). A database of 8 recordings from PhysioNet by using a neural network classifier were used and 93.39% accuracy was claimed to detect whether a patient is OSA positive or OSA negative. However, the accuracy achieved was low compared to our result and severity classification not included in the study. Another study extracted different linear and nonlinear features from SpO2 trained with 148 subjects and validated on two different databases (18). It gives an accuracy of 85.2% for data of 101 subject’s and an accuracy of 88.7% for other data from 71 subjects. However, the accuracy achieved in both cases was low compared to our result. Sleep apnea diagnosis based on real-time HRV analysis, which requires lab view, and used fuzzy expert system classifier was also proposed, but the high level of noise signal compromises the accuracy (16).
Only a few studies have been proposed for the detection of the apneic/hypopnea events on a minute-by-minute basis. In this sense, accuracy of 84.40% was obtained using 150 features and accuracy of 83.26% obtained with reduced 39 features from 25 subjects HRV and oximetry data from the University College Dublin (UCD) database where 25 sleep-disordered-breathing suspects (25). However, this work focuses only on detecting sleep apnea events and achieved low accuracy compared to our result. Another study has achieved an accuracy of 98.1% by using ECG and SpO2 for detecting sleep apnea which is less compared with our result and severity classification also not included compared with our result (19). Sleep apnea severity classification using the ECG signal alone has been proposed based on a deep learning approach (26). However, the method proposed works only for AHI <5 and AHI >30 and discriminates 5< AHI <30. In our method, the full range of AHI has been considered. In addition, we have shown that using the ECG signal alone is less accurate in detecting severity; 147 subjects were using ECG data for severity classification, claiming accuracy of 84.76%, specificity of 86.82 and sensitivity of 81.45% (12). However, in addition to its deteriorated accuracy, subjects representing mild and moderate OSA has not been considered.
Our proposed approach classifies apnea in minute-long segments and severity in hour-long segments. We also test our proposed method by using an additional local database. It performs well when compared with other methods in classification in a minute-by-minute apnea data. In addition, compared to other methods, which depend on binary severity classification, our method considers a wide range of AHI for severity level classification. Moreover, we have found that using ECG and SpO2 signals simultaneously offers benefits over using either signal alone in many ways. First, the combination is inherently more robust, as in the event of either channel is poor quality, the system can continue to make an analysis based on the other channel. Second, it improves the accuracy of using either of the signals alone. Third, periods of apnea-hypopnea can be directly linked to oxygen desaturations.
We acknowledge that the subjects with cardiac disorders, which was not considered in this paper, might have an impact on the reliability of the feature detection in ECG signal HRV.
Conclusions
This research presented a system for the automatic classification of simultaneously recorded ECG and SpO2 signals for subjects with sleep apnea. A model for automatic detection of sleep apnea using ECG signal alone and SpO2 signal alone was also presented and its effectiveness was evaluated. The sleep apnea detection and severity classification using the ECG signal was based on a selective set of RR-interval time-domain features that were fed to different classifiers including KNN, LDA and SVM, for classification. An overall accuracy of 99.1% and 88.89% were achieved for classifying apnea and the severity level, respectively. The results have demonstrated high performance and improved accuracy when using combined features from ECG and SpO2. The proposed model has a potential for adoption into clinical setting as a decision support system for physicians, especially for those in low resource setting where both the expertise and the means are in scarce.
Acknowledgments
The resources required for this research were provided by the school of Biomedical Engineering, Jimma institute of Technology, Jimma University and Jimma University Medical Center (JUMC). We would also like to thank Hallelujah General Hospital for allowing us collect data from their database.
Funding: None.
Footnote
Data Sharing Statement: Available at https://dx.doi.org/10.21037/ht-21-18
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/ht-21-18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. A written informed consent has been obtained from the participants prior to data collection. This research has been approved by Jimma University’s institutional review board (IRB: 36/2019) and the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stöwhas AC, Lichtblau M, Bloch KE. Obstruktives Schlafapnoe-Syndrom Praxis 2019;108:111-7. [Obstructive Sleep Apnea Syndrome]. [Crossref] [PubMed]
- Berry RB, Budhiraja R, Gottlieb DJ, et al. Rules for scoring respiratory events in sleep: update of the 2007 AASM Manual for the Scoring of Sleep and Associated Events. Deliberations of the Sleep Apnea Definitions Task Force of the American Academy of Sleep Medicine. J Clin Sleep Med 2012;8:597-619. [Crossref] [PubMed]
- Leppänen T, Kulkas A, Mervaala E, et al. Increase in Body Mass Index Decreases Duration of Apneas and Hypopneas in Obstructive Sleep Apnea. Respir Care 2019;64:77-84. [Crossref] [PubMed]
- Shamim-Uzzaman Q, Singh S, Chowdhuri S. Hypopnea definitions, determinants and dilemmas: a focused review. Sleep Science Practice 2018;2: [Crossref]
- Benjafield A, Valentine K, Ayas N, et al. Global Prevalence of Obstructive Sleep Apnea in Adults: Estimation Using Currently Available Data. Am J Respir Crit Care Med 2018;197:A3962.
- Memon J, Manganaro SN. Obstructive Sleep-disordered Breathing. In StatPearls. StatPearls Publishing. Bookshelf ID: NBK441909, 2021.
- Iannella G, Magliulo G, Lo Iacono CAM, et al. Positional Obstructive Sleep Apnea Syndrome in Elderly Patients. Int J Environ Res Public Health 2020;17:1120. [Crossref] [PubMed]
- Nwosu N. Obstructive Sleep Apnoea: Important things to know. BDJ Student 2019;26:40-2. [Crossref]
- Stearns JD, Stierer TL. Peri-operative identification of patients at risk for obstructive sleep apnea. Semin Anesth Perioper Med Pain 2007;26:73-82. [Crossref]
- Almazaydeh L, Elleithy KM, Faezipour M. A New Model for Diagnosing Sleep Apnea Through Features Extraction of the SpO2 Signal. UB Scholar Works, 2012;45.
- Almazaydeh L, Elleithy K, Faezipour M. Obstructive sleep apnea detection using SVM-based classification of ECG signal features. Annu Int Conf IEEE Eng Med Biol Soc 2012;2012:4938-41. [Crossref] [PubMed]
- Martín-González S, Navarro-Mesa JL, Juliá-Serdá G, et al. Heart rate variability feature selection in the presence of sleep apnea: An expert system for the characterization and detection of the disorder. Comput Biol Med 2017;91:47-58. [Crossref] [PubMed]
- Lin YY, Wu HT, Hsu CA, et al. Sleep Apnea Detection Based on Thoracic and Abdominal Movement Signals of Wearable Piezo-Electric Bands. IEEE J Biomed Health Inform 2016;21:1533-45. [Crossref] [PubMed]
- Ravelo-García A, Kraemer J, Navarro-Mesa J, et al. Oxygen Saturation and RR Intervals Feature Selection for Sleep Apnea Detection. Entropy 2015;17:2932-57. [Crossref]
- Al-Ratrout S, Hossen A. Support vector machine of wavelet packet spectral features for identification of obstructive sleep apnea. In: 2018 5th International Conference on Electrical and Electronic Engineering (ICEEE); 2018:380-3.
- M.S J. Robin CRR. Fuzzy based Expert system for sleep apnea diagnosis. Int J Eng Trends Technol 2016;35:555-8. [Crossref]
- Mostafa SS, Carvalho JP, Dias FM, et al. Optimization of sleep apnea detection using SpO2 and ANN. 2017 XXVI International Conference on Information, Communication and Automation Technologies (ICAT), 2017:1-6.
- Alvarez D, Hornero R, Marcos J, et al. Spectral analysis of electroencephalogram and oximetric signals in obstructive sleep apnea diagnosis. Annu Int Conf IEEE Eng Med Biol Soc 2009;2009:400-3. [Crossref] [PubMed]
- Punjabi M, Prabhu S. An ANN-Based Detection of Obstructive Sleep Apnea from Simultaneous ECG and SpO2 Recordings. In: Lecture Notes in Computational Vision and Biomechanics; 2019:603-13.
- Pan J, Tompkins WJ. A real-time QRS detection algorithm. IEEE Trans Biomed Eng 1985;32:230-6. [Crossref] [PubMed]
- Hastie T, Tibshirani R, Friedman J. Springer Series in Statistics. The Elements of Statistical Learning. Math Intell 2009;27:83-5.
- Bawa K, Sabherwal P. R-Peak Detection by Modified Pan-Tompkins Algorithm. Int J Adv Res Technol 2014;3:30-3.
- Durgabai RPL, Ravi Bhushan Y. Feature Selection using ReliefF Algorithm. IJARCCE 2014;3:8215-8. [Crossref]
- Khandoker AH, Palaniswami M, Karmakar CK. Support vector machines for automated recognition of obstructive sleep apnea syndrome from ECG recordings. IEEE Trans Inf Technol Biomed 2009;13:37-48. [Crossref] [PubMed]
- Xie B, Minn H. Real-time sleep apnea detection by classifier combination. IEEE Trans Inf Technol Biomed 2012;16:469-77. [Crossref] [PubMed]
- Banluesombatkul N, Rakthanmanon T, Wilaiprasitporn T. Single Channel ECG for Obstructive Sleep Apnea Severity Detection Using a Deep Learning Approach. TENCON 2018 - 2018 IEEE Region 10 Conference, 2018:2011-6.
Cite this article as: Ayalew MP, Nemomssa HD, Simegn GL. Sleep apnea syndrome detection and classification of severity level from ECG and SpO2 signals. Health Technol 2021;5:13.